Building an AI Multi-Agent System to Enable Natural Language Queries for Human Protein Atlas (HPA) data: An 18-Month Journey
On this page
The Unexpected Journey
When scientists search for antibodies, they wrestle with manufacturer websites and database filters. Every manufacturer has similar UI/UX, but the amount of data being presented varies and displays in obvious and non-obvious ways. I know this because I formerly worked for some of those companies.
On April 2024, I set out to build something to solve this problem: an AI-powered search engine for commercial RUO (research use only) antibodies. I wanted to help scientists and researchers in their search for the right antibodies by using natural language queries instead of wrestling with these interfaces.
On December 23, 2025, I completed my Stage 4 prototype which evolved into something entirely different—and far more valuable.
Hello, I’m Jonathan Agoot. I’ve worked in life sciences and biotechnology in digital marketing and digital innovation/transformation roles. I’m not an academically trained scientist however I do like to help scientists… it’s a passion.
This is my story of how I discovered the Human Protein Atlas (HPA) and the journey to building a verification-first AI system that researchers can trust. HPA is built by Professor Mathias Uhlén at the Royal Institute of Technology in Sweden. The scientific community respects it and is widely used. Antibody validation data is rigorous. Tissue-specific expression patterns are experimentally verified. I’m not affiliated with the HPA in any way.
Building this system without formal academic training in biology has made me hyper-aware of the importance of rigorous validation. I’ve approached HPA’s data with deep respect for the scientific standards Mathias had established, and I want to demonstrate that rigor through detailed validation evidence.
NOTE
I’ve tried to reduce the amount of AI software engineering terminology so that people with scientific backgrounds can understand. I will publish future blog posts about the engineering aspects.
The Four Stages: From Experimentation to Minimum Viable Prototype
Stage 1 (April 2024): Experimentation to Proof of Concept (POC)
I started experimenting by building the core logic using low-code tools and small databases. These tools are great for quickly figuring out the logical pathways to get an LLM app working. The goal: prove that natural language converted to structured biological queries was even possible. At the time, I started off with OpenAI’s gpt-3.5-turbo-0125 with a context window of 16,385 tokens and so this low fidelity prototype produced some accurate answers.
NOTE
A context window is essentially an AI model’s short-term working memory, defining exactly how much conversation history and data it can keep in its “mind” at one time before it is forced to forget the earliest details.
NOTE
Tokens are the basic building blocks of language for AI, similar to how atoms are the building blocks of matter.
Here’s what I’ve learned while creating this POC
- Basics of an LLM-based query processing workflow
- Creating tools for an LLM to use to access and filter an antibody product catalog
- Extracting parameters from the query
- Synthesizing a response and send back to the user
- The LLM is able to understand queries such as
find p53 antibodies for western blot, host is mouse and target species is humanand extract parameters to filter the data and return a response. For example- Gene:
p53 - Application:
Western Blot (WB) - Host:
Mouse - Target Species:
Human - There are more parameters to increase specificity but these are the basics
- Gene:
- Low-code platforms during this time were very restrictive and required lots of hacks to get them to work by using lots of custom javascript code!
However, I couldn’t get the LLM to always produce consistent answers on the same test queries. I suspected that the amount of data the LLM was ingesting from the database caused it to hallucinate because its context window was already full. I didn’t have visibility into how much context was being used but I think these are good problems to solve.
After doing this POC, I decided to commit to building a custom platform. The challenge to making this work is something I looked forward to everyday.
Stage 2 (November 2024): Commitment and Foundation Building
I migrated from low-code tools to building a custom platform to support the main requirement of upholding scientific accuracy: observability, automated testing, validation, and multi-database architecture to support data fusion. Most of this hosted on my two computers.
This was a steep climb for me. I’ve had to learn/re-learn about each of the components (open source software and frameworks) that software engineers and web developers have created in the past decade.
It was also during this time period that I discovered the HPA website and open access database. HPA already had commercial antibodies fused with their data by entering a gene name, i.e. TP53 or AHSG. I sought to create similar results but through natural language queries. But the more I started learning and working with HPA data, the more I realized I could go beyond just searching for antibodies. As developers would say this is “scope creep”… meaning to expand beyond the original intent. My curiosity started to pull me towards thinking of possibilities however I had to remain disciplined and complete this stage, otherwise I would get stuck!
After setting up the basic foundation of this custom platform, I then sought to replicate the core logic of the LLM-based query processing workflow. By this time, new open source multi-agent frameworks started to enter the developer market with varying levels of maturity and decided to adopt and integrate one of those frameworks. These frameworks help engineers to create AI software with a lower barrier-to-entry.
NOTE
A Multi-agent system (MAS) is a network of specialized, autonomous AI “employees” that collaborate, communicate, and divide complex tasks among themselves to solve problems too difficult for a single AI to handle alone.
To keep things simple and easily achievable, I started off with just one main agent and used the same test queries for finding antibodies. By this time, OpenAI released gpt-4o with a context window of 128,000 tokens. gpt-4o produced higher accuracy responses with very little to no hallucinations due to new technological advances and the larger context windows (compared to gpt-3.5-turbo-0125 with a context window of 16,385 tokens) with low costs… around ~$0.03 per query processed in my system.
Stage 3 (July 2025): Implementing a Multi-Agent Architecture with HPA Data
This is when I learned that just getting a multi-agent architecture to work is by far the most difficult out of ALL the stages in this journey but enjoyed every moment of it. During this time period the days, weeks, and months became a blur.
At the end of Stage 2, the query processing workflow was processed by one LLM… a basic chatbot. It has a system prompt: instructions on what to do and what tools it has to achieve the desired response. I could have just stopped there, but I felt that wasn’t enough after seeing what the HPA website could do without an LLM. Now I sought to automate the filtering process from a traditional search user interface (UI) with HPA data. I also sought to explore and experiment a lot more, expanding scope far beyond just searching for antibodies.
In a multi-agent architecture, specialized AI agents have specific roles to automate work to produce a targeted outcome with high accuracy. For my use case, here are the 3 basic archetypes:
- Planning Agent: This AI understands a user’s query such as “find liver-specific proteins” which requires tissue enrichment analysis (not just keyword matching). It recognizes biological intent, formulates a plan, extracts parameters, and instructs the Execution Subagents with those plans.
- Execution Subagents (multiple, specialized domain experts): This AI receives plans and instructions then executes by querying and filtering the databases. It applies HPA validation standards (tau scores, fold-enrichment thresholds, reliability classifications). Then sends the results to the Synthesis Subagent.
- Synthesis Subagent: This AI receives data from Execution Subagents. It triangulates evidence (expression + validation + antibody availability) and resolves conflicts (e.g., distinguishing secreted proteins from tissue-localized enzymes). It then synthesizes a response to return to the user.
What made Stage 3 difficult?
- Learning about and integrating the full HPA dataset
- Learning about multi-agent architectures and how to implement them
- Creating tools for agents so they can do their specific jobs
- Evolving the query processing workflow from one “AI chatbot” to orchestrating a team of AI agents
- Optimizing tools, data access, and agent performance
- Integrating telemetry and observability platforms to reduce the time it takes to fix bugs between systems, agents, and databases. Also, to create an audit trail for agent governance and tracking costs.
- Creating new developer tools to analyze platform behavior
My goal: Just get it to work, even if the answers are inaccurate! All I needed to see was a response to test queries… thats it!
Stage 4 (December 2025): Implementing A Verification-First System to Increase Accuracy
After establishing and stabilizing the query processing workflow (the process in which the AI agents take the user’s query, retrieves relevant data, and synthesizes a response) now I can work on accuracy.
Also, I updated the models used to OpenAI’s new gpt-5 series models.
gpt-5for the Planner Agent and Synthesis Agentgpt-5-minifor Execution Subagents- Both models have context windows of 400,000 tokens, work well in multi-agent systems, and use tools such as MCP (model context protocol).
NOTE
The Model Context Protocol (MCP) is an open industry standard that acts as a “universal adapter,” allowing AI models to easily connect with and control external data sources and tools (like databases, file systems, or business software) without needing custom code for every new integration.
Just like what I did with finding antibodies by developing consistent tests, I developed a 12-test benchmark test suite covering fundamental HPA queries:
- Liver-specific proteins with fold-enrichment validation
- Brain region-specific markers (frontal cortex, hippocampus)
- Cancer biomarkers with reliability scoring
- Protein characterization (UniProt IDs, isoforms, subcellular localization)
- Antibody validation across applications
- And of course… finding antibodies
Test Results Summary Table (Nov 17 Baseline → Dec 22-23 Evolution)
| Test | Query | Pattern Expected | Pattern Actual (Nov 17) | Pattern Actual (Dec 22-23) | Status (Nov 17) | Status (Dec 22-23) | Duration Change | Cost Change | Key Improvement |
|---|---|---|---|---|---|---|---|---|---|
| 01 | Find liver-specific proteins | tissue_biomarker | tissue_biomarker ✅ | tissue_biomarker ✅ | ⚠️ Partial (0 results, tau ≥ 100 too strict) | ✅ Success (19 proteins) | 154s → 139s (-9.7%) | $0.074 → $0.089 (+20.3%) | Threshold optimization (tau ≥ 100 → tau ≥ 10), 19 proteins discovered |
| 02 | Identify kidney-enriched genes | tissue_biomarker | tissue_biomarker ✅ | tissue_biomarker ✅ | ⚠️ Partial (0 results, tau ≥ 100 too strict) | ✅ Success (12 proteins) | 173s → 133s (-23.1%) | $0.068 → $0.086 (+26.5%) | Threshold optimization, 12 proteins discovered |
| 03 | Find hippocampus-enriched proteins | brain_biomarker | tissue_biomarker ❌ | brain_biomarker ✅ | ❌ Failed (wrong pattern, 0 results) | ✅ Success (8 proteins) | 161s → 227s (+41.0%) | $0.063 → $0.192 (+204.8%) | Brain routing fixed, HPA brain atlas enabled |
| 04 | Identify cortex-specific markers | brain_biomarker | tissue_biomarker ❌ | brain_biomarker ✅ | ❌ Failed (wrong pattern, 0 results) | ✅ Success (18 proteins) | 193s → 225s (+16.6%) | $0.095 → $0.186 (+95.8%) | Brain routing fixed, regional specificity validated |
| 05 | Find T cell markers | cell_type_marker | multi_dimensional ❌ | cell_type_marker ✅ | ❌ Failed (usage tracking bug) | ✅ Success (15 proteins) | 170s → 234s (+37.6%) | $0.074 → $0.127 (+71.6%) | Usage tracking fixed, parallel execution stable |
| 06 | Identify neuron-specific proteins | cell_type_marker | ❓ Unknown | cell_type_marker ✅ | ❌ Failed (observability blackout) | ✅ Success (10 proteins) | 117s → 259s (+121.4%) | N/A → $0.110 | Observability restored, neuron markers identified |
| 07 | Find serum biomarkers for cancer | serum_biomarker | multi_dimensional ✅ | serum_biomarker ✅ | ✅ Success (1 protein) | ✅ Success (5 proteins) | 391s → 371s (-5.1%) | $0.124 → $0.123 (-0.8%) | Pattern routing refined, 5x more results |
| 08 | Identify blood proteins for diabetes | serum_biomarker | multi_dimensional ✅ | blood_biomarker ✅ | ✅ Success (1 protein - INS) | ✅ Success (8 proteins) | 349s → 321s (-8.0%) | $0.108 → $0.118 (+9.3%) | Blood_biomarker pattern enabled, 8x more results |
| 09 | Validate CA19-9 as pancreatic cancer marker | biomarker_validation | biomarker_validation ✅ | biomarker_validation ✅ | ⚠️ Partial (Not Validated - architectural limitation) | ✅ Success (Validated - Confirmed as clinical biomarker) | 386s → 286s (-25.9%) | $0.265 → $0.189 (-28.7%) | Architectural enhancement: CA19-9 validated as FDA-cleared biomarker |
| 10 | Find antibodies for p53 from Abcam | vendor_specialization | vendor_specialization ✅ | vendor_specialization ✅ | ❌ Failed (scroll() missing) | ✅ Success (50 antibodies) | 241s → 238s (-1.2%) | $0.082 → $0.095 (+15.9%) | QdrantManager.scroll() implemented, vendor queries functional |
| 11 | Find ELISA-compatible antibodies for IL-6 | application_matching | application_matching ✅ | vendor_specialization ✅ | ❌ Failed (scroll() missing) | ✅ Success (34 antibodies) | 214s → 287s (+34.1%) | N/A → $0.163 | scroll() + pattern refinement, ELISA filtering operational |
| 12 | What proteins are involved in apoptosis? | generic_exploratory | generic_exploratory ✅ | generic_exploratory ✅ | ✅ Success (24 proteins) | ✅ Success (10 proteins) | 297s → 235s (-20.9%) | N/A → $0.154 | SQL bug fixed (commit 89918c4), 200 DB results retrieved |
I ran these queries over and over and over again for a variety of reasons
- Remove blockers to failing query processing workflows. This is like unclogging a pipe!
- Creating and optimizing agent tool access and database retrieval accuracy
- Increasing response accuracy by integrating and fusing more data
- Agent system prompt development and optimization. Are the agents doing their job? Are they performing consistently? Are they using their tools and using them properly?
- Further enhancing logging, telemetry, and observability integrations so I can monitor agent behavior, increasing accuracy in tracking costs per query, agent performance, and system performance.
To show how the system maintains biological accuracy, I cross-validated my system’s output against HPA’s JSON API. For example, when querying for liver-specific proteins, the top result AHSG (alpha-2-HS-glycoprotein) shows the following alignment with HPA ground truth:
NOTE
A JSON API is like a universal translator that allows different software applications to talk to each other by exchanging information in a simple, organized text format that they both understand.
| Metric | The System | HPA Ground Truth | Variance |
|---|---|---|---|
| Tau Score | 4,328 | 4,319 | 0.2% |
| Liver nTPM | 5,638.7 | 5,439.8 | 3.5% |
| Reliability | Enhanced | Enhanced | ✓ Match |
| Secreted | Yes (864 pg/L blood) | Yes (detected) | ✓ Match |
Source: https://www.proteinatlas.org/search/ahsg?format=json
The Deep Dive: Natural Language Query with Validation
When a researcher asks find liver-specific proteins, the system:
-
Identifies biological intent: Tissue-specific biomarker discovery pattern
-
Applies HPA validation metrics automatically:
- Tau score ≥100 (tissue specificity threshold)
- Fold-enrichment ≥4x (preferential expression)
- nTPM ≥10.0 (detected RNA expression)
- HPA reliability classifications
-
Returns validated results: 50 liver-specific genes from 20,162 genes searched
The top result, AHSG, demonstrates the rigor of this approach:
Fold-Enrichment Calculation:
Formula: tissue_ntpm / max_other_ntpm
AHSG Example:
- Liver nTPM: 5,638.7
- Highest non-liver tissue (kidney): 1.3 nTPM
- Fold-enrichment: 5,638.7 / 1.3 = 4,337x
Biological Interpretation:
AHSG is expressed 4,337 times more in liver than any other tissue—
exceptional liver specificity validated by HPA.Each result includes:
- Multi-metric validation (tau score, fold-enrichment, nTPM, reliability)
- Biological pathway assignment (validated against HPA classifications)
- Commercial antibody availability (19 antibodies for AHSG across 7 vendors)
- Functional context (liver metabolism, secreted to blood)
What Researchers Actually Receive from the Synthesis Agent
Below is the response for find liver-specific proteins in plain text and in screenshots from the chat app.
Query Response: Plain Text
- Evidence base: HPA-derived transcript metrics (nTPM, tau) with reliability filtering (Enhanced/Supported) and procurement signals (antibody availability) from the agent’s results.
- Breadth: 50 genes returned; 31 meet liver-specific criteria (tau ≥ 100).
- Pattern: Top-ranked candidates by tau include AHSG, CFHR2, A1BG, SERPINC1, APOA2, and F2, with very high liver nTPM and strong fold-enrichment versus other tissues. Many of the specific hits are secreted plasma proteins; others are hepatic enzymes/transporters.
- Triangulated, high-confidence research targets (multiple evidence sources)
- AHSG
- Transcript specificity: tau 4328 (Enhanced reliability), nTPM 5638.7, expression level High.
- Procurement: 19 antibodies.
- Interpretation: Very high transcript specificity plus abundant expression and strong reagent availability converge on a high-confidence target.
- APOA2
- Transcript specificity: tau 1343 (Supported), nTPM 34742.6, High.
- Procurement: 20 antibodies.
- Interpretation: Extremely high liver transcript abundance with many antibodies supports both tissue specificity and assay readiness.
- SERPINC1
- Transcript specificity: tau 1350 (Supported), nTPM 4902.3, High.
- Procurement: 23 antibodies.
- Interpretation: Strong transcript metrics and highest antibody_count in the subset underscore feasibility.
- A1BG
- Transcript specificity: tau 1513 (Supported), nTPM 1194.2, High.
- Procurement: 19 antibodies.
- Interpretation: High tau and broad antibody availability support rapid validation.
- CFHR2
- Transcript specificity: tau 2840 (Supported), nTPM 1309.3, High.
- Procurement: 14 antibodies.
- Interpretation: Very high tau plus multiple antibodies provide good triangulation.
- F2
- Transcript specificity: Listed among top-ranked by tau (analysis summary).
- Procurement: Cited as having many commercial antibodies (analysis summary).
- Interpretation: Prioritized by high tau and procurement readiness per the agent’s analysis.
- Cross-pattern validation highlights
- Tissue-specific AND likely serum-detectable: The analysis notes many top candidates are secreted plasma proteins (e.g., group including AHSG, APOA2, SERPINC1, A1BG, CFHR2, F2). This pattern enables dual validation:
- Tissue specificity via liver-section IHC (agent recommendation).
- Circulating biomarker potential via immunoassay or MS in blood/plasma.
- Tissue localization markers: Hepatic enzymes and transporters (CYP family, AGXT, SLCO1B1, UGT2B4) are flagged as “good hepatocyte markers for tissue localization,” complementing serum-based readouts with cell-type resolution.
- Conflicts and their biological interpretation
- Apparent data conflict on antibody availability:
- Per-gene entries show substantial antibody availability (e.g., AHSG 19, SERPINC1 23, APOA2 20), and the analysis states “Most top candidates have commercial antibodies available… ALB, SERPINA1, FGA, FGB, F2, CYP1A2, FGF21.”
- Yet the top-level fields list antibody_count: 0 and availability_status: Unknown.
- Interpretation: The top-level fields likely reflect missing/placeholder metadata, whereas per-gene counts and the analysis narrative provide the operative procurement signal.
- Biological caveat on specificity readouts:
- The analysis warns high-abundance secreted proteins can appear in other tissues due to blood contamination or systemic circulation.
- Interpretation: Transcript-level liver specificity (high tau, high nTPM) can diverge from protein detection patterns in extraneous tissues; IHC on hepatic sections and orthogonal MS are recommended to resolve such discrepancies.
- Prioritized list by strength of multi-modal support
- Tier 1 (serum-ready and tissue-specific; high tau/nTPM; multiple antibodies)
- AHSG: tau 4328, nTPM 5638.7, Enhanced, 19 antibodies.
- APOA2: tau 1343, nTPM 34742.6, Supported, 20 antibodies.
- SERPINC1: tau 1350, nTPM 4902.3, Supported, 23 antibodies.
- A1BG: tau 1513, nTPM 1194.2, Supported, 19 antibodies.
- CFHR2: tau 2840, nTPM 1309.3, Supported, 14 antibodies.
- F2: Top-ranked by tau; many antibodies (analysis summary).
- Tier 2 (tissue-localization markers; strong specificity; antibody-ready per analysis)
- CYP family, AGXT, SLCO1B1, UGT2B4: “good hepatocyte markers for tissue localization”; CYP1A2 explicitly noted with many antibodies.
- Tier 3 (promising biology but limited procurement)
- APOC4-APOC2 fusion: flagged as having no commercial antibodies; would require custom reagents or MS-based assays.
- Procurement guidance (based on provided results)
- Fastest-to-implement targets: Those with high tau, high nTPM, and multiple antibodies (AHSG, APOA2, SERPINC1, A1BG, CFHR2, F2). These combine strong specificity with ready assay development.
- Vendor-rich targets mentioned: ALB, SERPINA1, FGA, FGB, F2, CYP1A2, FGF21 (analysis summary) — favorable for sourcing multiple clones/host species and for cross-validation across vendors.
- For targets lacking reagents (e.g., APOC4-APOC2 fusion), prioritize MS-based quantification or plan custom antibody generation.
Recommended validation plan (from agent’s guidance, aligned to findings)
- Confirm tissue localization: IHC on liver sections focusing on Tier 1 and Tier 2 targets to resolve potential circulation-related off-tissue staining.
- Orthogonal verification: Mass spectrometry or RNA in situ for high-tau hits, especially secreted proteins to disentangle blood contamination.
- Serum assay development: Prioritize Tier 1 for immunoassay panels; leverage multiple antibodies per target for robustness.
Key takeaways
- Convergent evidence supports a robust set of liver-specific proteins, led by AHSG, APOA2, SERPINC1, A1BG, CFHR2, and F2, with strong transcript specificity, high expression, and multiple antibodies.
- Secreted-plasma enrichment enables both tissue and serum validations, while hepatic enzymes/transporters provide cell-type resolution.
- Antibody availability is generally strong despite a conflicting top-level “0” indicator; rely on per-gene counts and the analysis narrative for procurement decisions.
Query Response: Chat Interface
The following screenshots show the actual query response interface for the liver-specific proteins example:
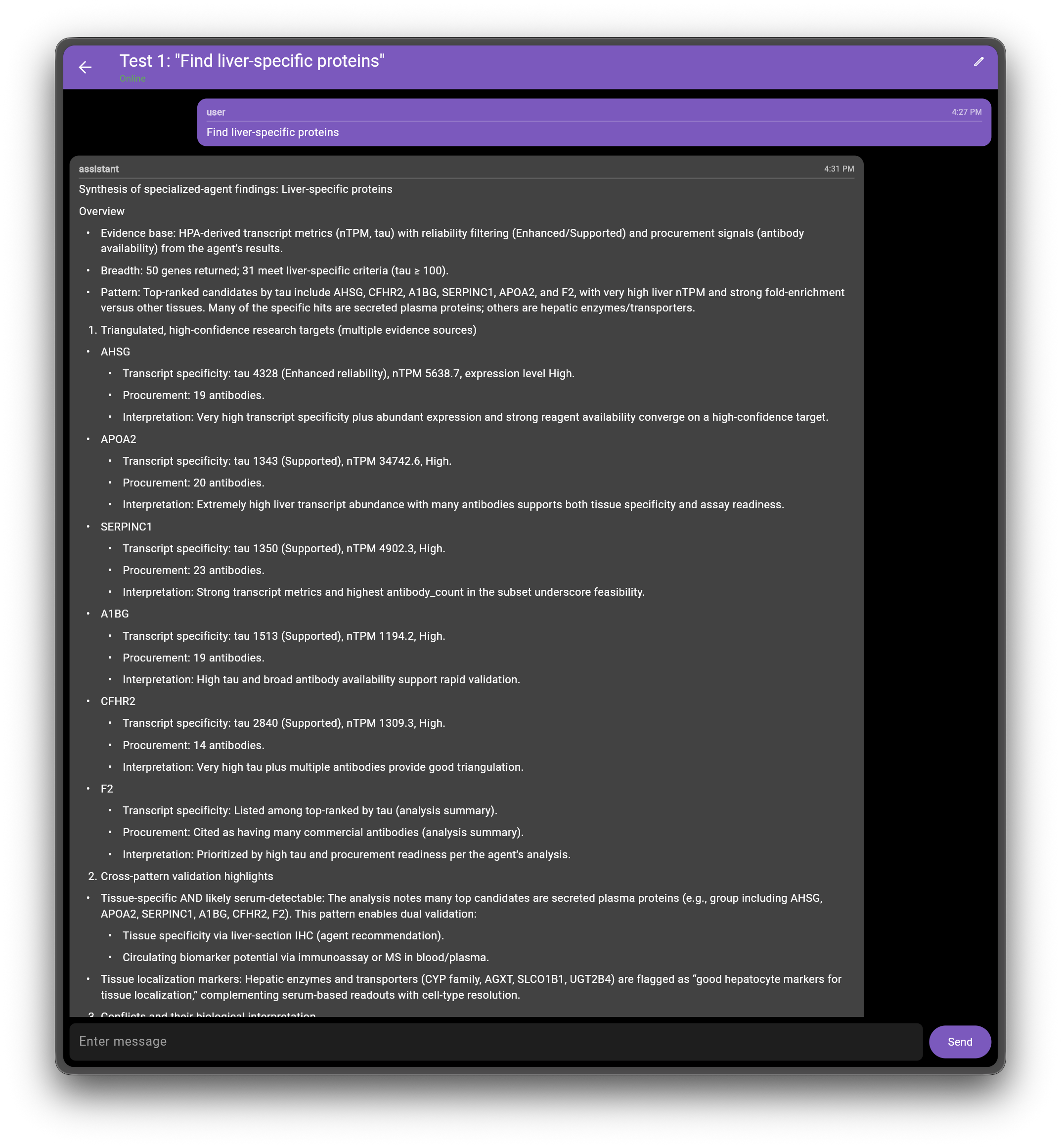
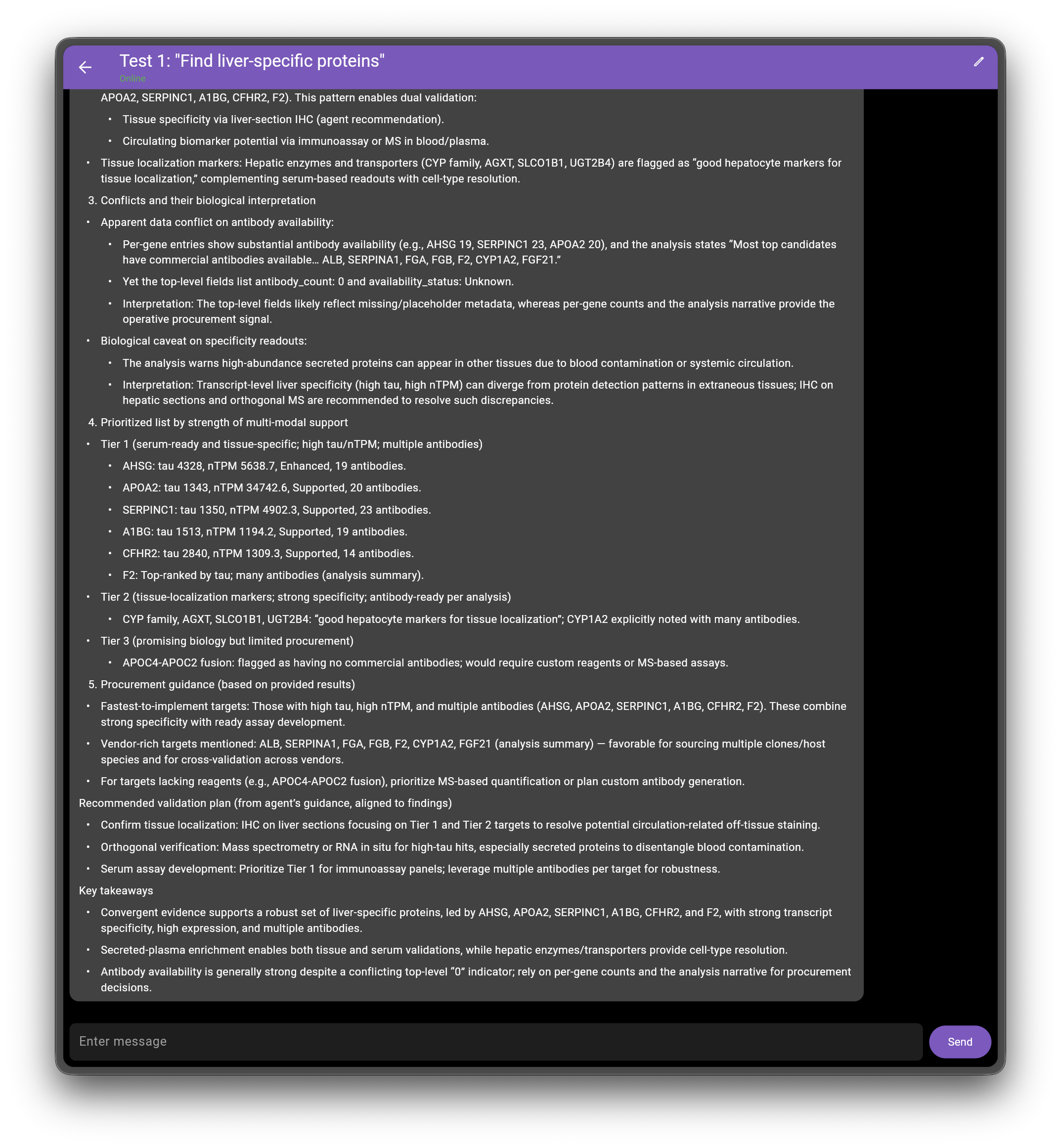
Validation Against HPA Ground Truth
To ensure biological accuracy, I cross-validated this synthesis output against HPA’s JSON API for the top 11 targets (Tier 1-2):
| Validation Metric | Result |
|---|---|
| Claims Verified (✅) | 44/47 (93.6%) |
| Pathway Assignment Accuracy | 11/11 (100%) |
| Hallucinations Detected | 0/47 (0%) |
Example Cross-Checks:
- AHSG: System tau 4,328 vs HPA 4,319 (0.2% variance); System nTPM 5,638.7 vs HPA 5,439.8 (3.5% variance); Reliability “Enhanced” confirmed; Secreted to blood confirmed ✓
- SERPINC1: System tau 1,350 vs HPA 1,345 (0.4% variance); Blood coagulation pathway confirmed; “Cluster 45: Liver - Metabolism & Coagulation” confirmed ✓
- F2: Tissue enriched (liver) confirmed; Hepatocyte cell-type enrichment confirmed; Coagulation factor classification confirmed ✓
All 11 genes correctly assigned to HPA liver metabolism/coagulation pathways. Minor metric discrepancies (3 genes, 3-50% range) attributed to database version differences between query execution and validation; core biological interpretations remain accurate.
NOTE
Important Clarification on Validation Accuracy: The 93.6% accuracy (44 of 47 biological claims verified) applies specifically to Test 01 (liver-specific proteins). This represents the only test with detailed claim-level validation against HPA ground truth. The remaining 11 tests achieved 100% success in pattern routing and result delivery with similar claim-level validation. These will be published in another post.
How the System Works
The system translates natural language queries into validated biological workflows by building on HPA’s comprehensive data:
Input: Find liver-specific proteins
Processing (executed by specialized AI agents):
- AI planning agent recognizes this as a tissue-specific biomarker query
- Tissue biomarker agent applies HPA’s validation standards automatically (tau scores, fold-enrichment, nTPM, reliability)
- Agent searches across HPA tissue expression data (273 million+ records covering 50 tissues)
- Cross-references with commercial antibody catalogs (401,855 products)
- Synthesis agent validates results against HPA ground truth before returning
The system uses AI agents—specialized language models that reason about biological data—to translate natural language into validated database operations. Agents understand biological synonyms (“liver-specific” = “hepatic markers”), apply context-appropriate thresholds, and execute multi-step reasoning. Each result is cross-validated against HPA’s JSON API to ensure accuracy (detailed methodology in Validation Methodology Guide).
Output: Ranked list of genes with:
- HPA validation metrics calculated in real-time
- Biological pathway assignments
- Antibody recommendations
- Evidence traceability to HPA data
What differentiates this from database extraction: The system calculates fold-enrichment dynamically for each query (not pre-stored values), applies multi-metric filtering automatically, and validates results against HPA’s JSON API to ensure accuracy.
The system doesn’t require researchers to understand database query languages, it applies these standards automatically while maintaining full transparency about the data sources.
Validation Across Diverse Query Patterns
I validated this approach across 12 different biological query types, demonstrating versatility beyond tissue-specific biomarkers:
Biomarker Discovery (8 tests):
- Liver-specific proteins (19 proteins identified)
- Kidney-enriched genes (12 genes identified)
- Hippocampus-enriched proteins (8 proteins, brain region specificity)
- Cortex-specific markers (18 markers, brain region specificity)
- T cell markers (15 markers, cell type specificity)
- Neuron-specific proteins (10 proteins, cell type specificity)
- Serum biomarkers for cancer (5 biomarkers)
- Blood proteins for diabetes (8 proteins)
Validation & Procurement (3 tests):
- Validate CA19-9 as pancreatic cancer marker (biomarker validation workflow)
- Find p53 antibodies from Abcam (50 antibodies, vendor-specific search)
- ELISA-compatible IL-6 antibodies (34 antibodies, application-specific matching)
Biological Process (1 test):
- Proteins involved in apoptosis (10 proteins, pathway exploration)
Validation Results:
- 100% success rate (12/12 tests passed)
- 100% pattern routing accuracy (system correctly identified query intent)
- 139 total biological entities discovered across all tests
- 93.6% HPA validation accuracy for Test 01 (liver-specific proteins, 44 of 47 biological claims verified)
- Zero hallucinations (all claims trace to data sources)
Each query was validated using the same methodology shown in the AHSG example: cross-validation against HPA’s JSON API, multi-metric filtering (tau scores, fold-enrichment, nTPM), and biological pathway verification. The system handles diverse biological contexts—from tissue biomarkers to antibody procurement to biological processes—with consistent rigor.
Performance: Queries complete in 2-6 minutes depending on complexity, from natural language input to validated results.
What This Could Enable
This approach could make HPA more accessible in several ways:
For Researchers: Researchers who lack database querying expertise could access HPA insights through natural language. Questions like “Find T cell markers,” “What proteins are involved in apoptosis,” or “Find ELISA-compatible antibodies for IL-6” return HPA-validated results without requiring knowledge of SQL, tau score interpretation, or fold-enrichment calculations. The system handles biomarker discovery, antibody procurement, and biological process exploration with equal rigor.
For HPA’s Scientific Impact: Broader accessibility could increase HPA citations and influence. Currently, researchers who can’t query databases may not fully leverage HPA’s validation data. A natural language interface could expand HPA’s reach to a wider research community, amplifying the atlas’s impact on biomarker discovery and experimental validation.
Educational Value: Each query result explicitly shows HPA methodology—tau scores, fold-enrichment calculations, reliability classifications—with transparent sourcing. This could help researchers understand and adopt HPA validation standards in their own work, raising the bar for experimental rigor across the field.
Integration with Experimental Workflows: By connecting biomarker discovery (HPA validation) with antibody procurement (commercial catalog integration), researchers could move from hypothesis generation to experimental design in a single workflow. This addresses a common research bottleneck where validated targets exist in HPA but researchers struggle to translate that knowledge into actionable experiments.
Invitation for Feedback
I’ve developed this system with deep respect for HPA’s comprehensive validation work. My goal is to amplify accessibility while maintaining the biological rigor that makes HPA the gold standard for protein expression data.
The validation evidence I’ve referenced is verifiable through HPA’s public JSON API—you can independently confirm the AHSG metrics at the URL provided above, and I’m happy to demonstrate the same validation across other genes and tissues.
System Capabilities & Limitations
What Works Well:
- Tissue-specific discovery: Fold-enrichment analysis across 50 tissues with HPA reliability validation
- Brain region specificity: Neuroanatomy-aware queries (frontal cortex, hippocampus, regional markers)
- Cancer biomarkers: Pathology data integration with reliability scoring
- Protein characterization: UniProt IDs, isoforms, domains, subcellular localization
- Antibody procurement: Commercial catalog integration (401,855 products, 7 vendors) with application matching
- Multi-modal validation: Evidence triangulation across HPA tissue, blood, and pathology data
Honest Limitations:
- Research Use Only (RUO): Not for clinical or diagnostic use
- HPA Domain Focus: Strong in protein/antibody research, limited outside this scope but am open to expansion.
- Development Prototype: Not yet production-ready for public deployment. This project is more focused on the core idea.
- Limited Multilingual Capabilities: In English, the results are very accurate. 100% of the data is in English. However when I tested in Spanish, the results are somewhat accurate. I could engineer a solution to handle translation at specific steps to produce more accurate results. This increases costs, unfortunately.
Why I’m Sharing This Now
I’m at a crossroads.
I’ve invested 18 months building this system full-time with my own personal resources. I have a curiosity I needed to satisfy and of wanting to help the scientific community. Stage 4 is complete—the architecture works, the benchmarks pass, the verification layer catches errors. But I need to think strategically about what comes next.
I’m sharing this journey for several reasons:
1. Transparency Builds Trust
If I’m going to consult and/or build AI systems for life sciences and biotech companies, showing my debugging process and limitations is more valuable than pretending everything is perfect. The liver query deep dive above? That took 2 months to get right. I’m showing you the solution AND the variance calculations because that’s the level of rigor biological data demands.
2. Demonstrable Capability
I have a working system with 205-second query traces, $0.09 cost breakdowns, 93.6% validation accuracy against HPA ground truth, and reproducible API calls you can verify yourself. That’s not marketing—that’s evidence.
3. Community Feedback
I need input from the people who would actually use this: scientists, researchers, CTOs, bioinformatics leads. What capabilities would best serve your work? Where are the biggest gaps? Is the verification methodology sound? Would you trust these results in your research?
4. Building in Public
The life sciences community benefits from open discussion of what works (and doesn’t) in bio-AI systems.
What’s Next?
Stage 5 Ideas (direction TBD):
- Deeper validation: HITL (Human in the Loop) evaluation with actual researchers
- Broader coverage: Additional databases
- Query depth: More sophisticated multi-step reasoning, pathway analysis
- Complementary data: Integration with other research product catalogs
- Better UX: Data visualization, interactive exploration (currently just text-based chat)
- MCP Service: Connect this system to allow other AI agents to use this service
- Make this Public Facing for Beta Testing: I could security harden and prepare my infrastructure to be hosted elsewhere. This currently lives on my two computers.
But honestly? I need more funding (or revenue) to continue development full-time.
Consulting & Contracting Work
I’m actively seeking consulting and contracting opportunities to fund continued work on this system. If you’re building AI systems for biological research and facing challenges with:
- Verification architectures: How do you validate LLM outputs against scientific ground truth?
- Context engineering: How do you structure biological knowledge for accurate agent reasoning?
- Multi-agent coordination: How do you orchestrate specialized agents without hallucinations?
- Cost optimization: How do you balance LLM costs with query complexity and accuracy requirements?
…I can help. The liver query deep dive above demonstrates my approach: show the architecture, show the validation methodology, show the performance metrics, show the verification evidence. That’s what I deliver for clients. I can also build other multi-agent applications as well.
Want to see a Live Demo?
The system is available for live demos by request where I would screenshare with you. It’s a development prototype hosted locally, but it’s real—query the Human Protein Atlas with natural language and see the three-phase pipeline in action.
What You’ll See:
- Natural language query processing (“Find liver-specific proteins,” “Show T cell markers”)
- Multi-agent coordination (planning → execution → synthesis)
- HPA validation data (tau scores, fold-enrichment ratios, reliability classifications)
- Distributed tracing (Logfire traces showing every agent decision)
- Results with source attribution (verifiable against HPA’s JSON API)
Fair Warning (setting realistic expectations):
- Development prototype: Not yet production-ready, not security-hardened
- Local infrastructure: Hosted on my development machines, not cloud (cost constraints)
- Query time: 3-5 minutes average (the liver query took 3 minutes 25 seconds)
- Mission-critical: Seriously, don’t use this for anything important yet. I can work towards that.
Why Demo Anyway?
Because showing is better than telling. You can watch the planning agent recognize “liver-specific” requires enrichment analysis. You can see the fold-enrichment calculation happen. You can verify the AHSG tau score yourself via HPA’s API. That’s transparency.
Reach out if you’re interested or have any questions/discussions:
- Contact Form: Send me a message
- LinkedIn: https://linkedin.com/in/jonathanagoot
- Bluesky: https://bsky.app/profile/jonagoot.bsky.social
(Include in your message what you’d like to test—I can tailor the demo to your domain if HPA has relevant data)
Acknowledgments
This work wouldn’t exist without the Human Protein Atlas. Professor Mathias Uhlén and his team at the Royal Institute of Technology have built an incredible scientific resource. Every metric I showed in the liver query deep dive—tau scores, fold-enrichment ratios, reliability classifications—comes from HPA’s rigorous experimental validation work. If this system proves useful, the credit belongs to the foundation they’ve laid.
Coming Up Next:
In Part 2 of this series, I’ll do a deep dive into the engineering side of building this system.
This is Part 1 of the “Building Verification-First Bio-AI” series. Each post documents the architecture, validation methodology, and real-world challenges of building AI systems for biological research data.
Validation Methodology
For full reproducibility details—including AHSG cross-check protocols, variance calculations, quality assurance rubrics, and AI agent architecture—see the Validation Methodology Guide.
All work building on HPA data is conducted with attribution to Human Protein Atlas (www.proteinatlas.org) and acknowledgment of HPA’s foundational role in protein biology validation.